CURE
Unpacking protein structure-to-function relationships
through large, high-resolution, quantitative datasets.
OUR MISSION
Innovative Protein Engineering
The Design-to-Data workflow was developed in the Siegel Lab with the central research of improving the current predictive limitations of protein modeling software by functionally characterizing single amino acid mutants in a robust model system. This workflow is undergraduate-friendly, and students have an opportunity to practice protein design, kunkel mutagenesis, and enzyme characterization assays. The workflow is intuitively organized through engineering’s conceptual progression of design-build-test.

HOW IT WORKS
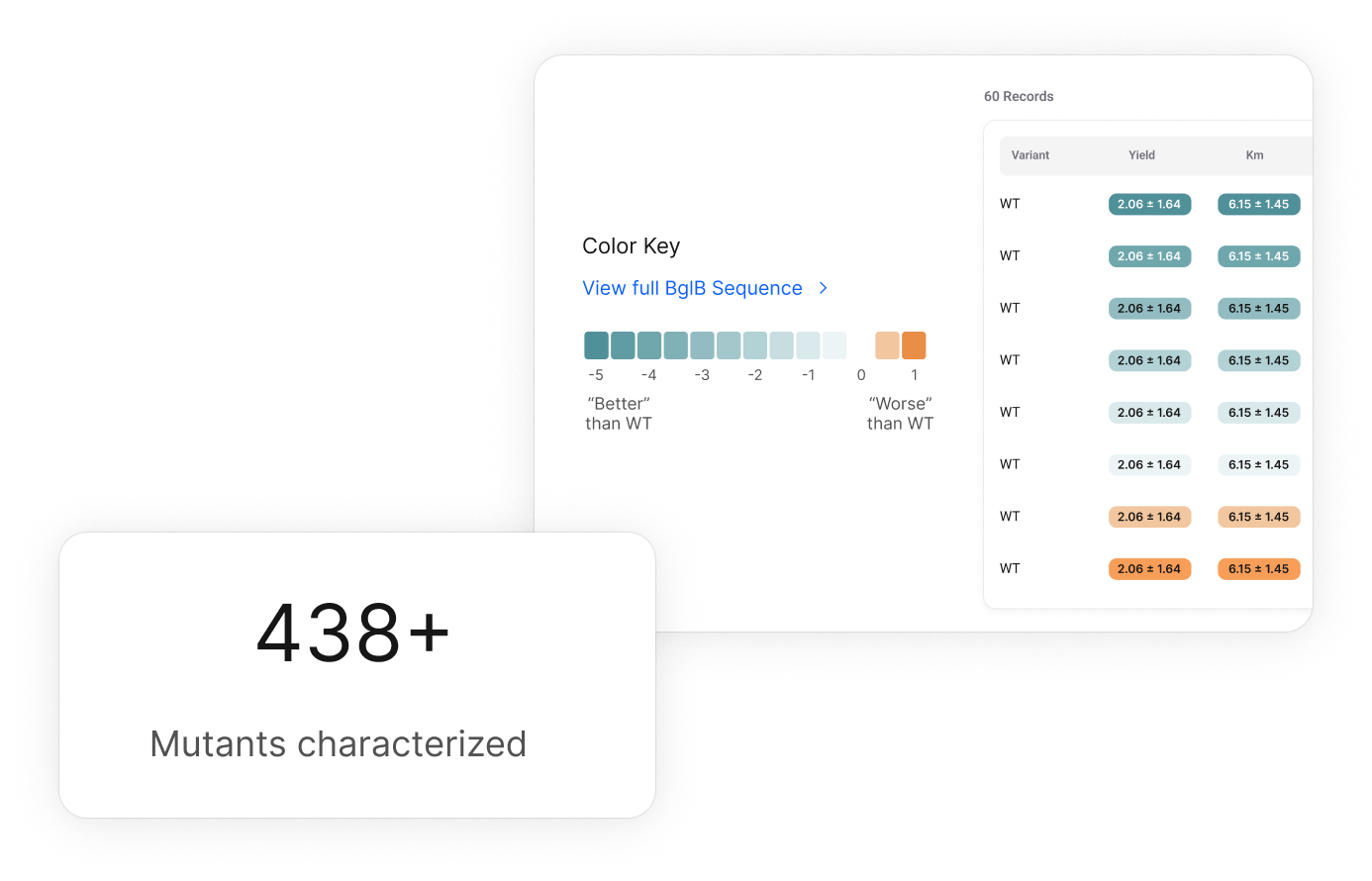
Analyze and Submit
D2D students upload their colorimetric kinetic and thermal assay data for enzyme va that they studied.
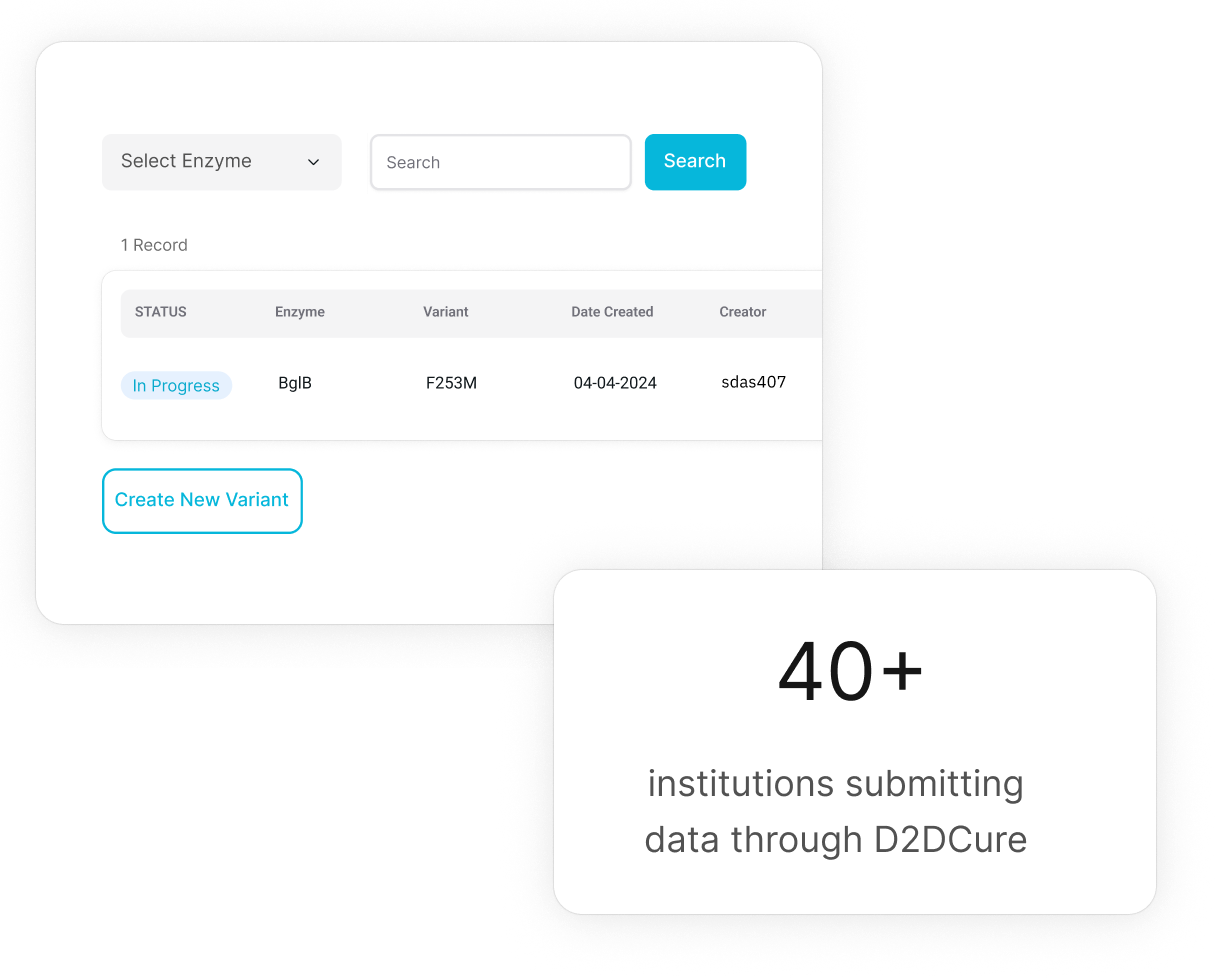
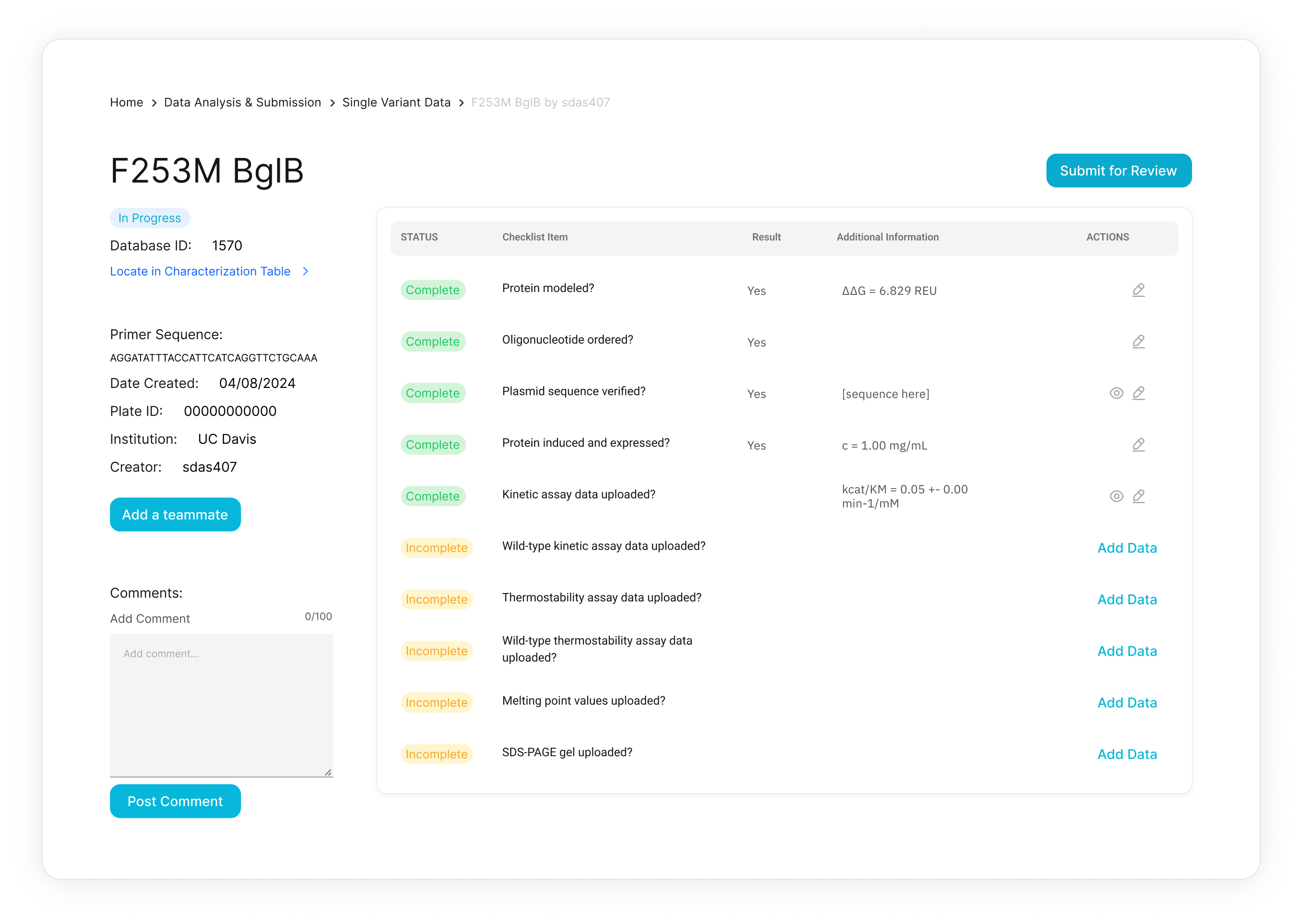
Curate
D2D faculty and admin in a two-step process review and approve data with appropriate controls; data that fails to meet network standards is flagged for replication.
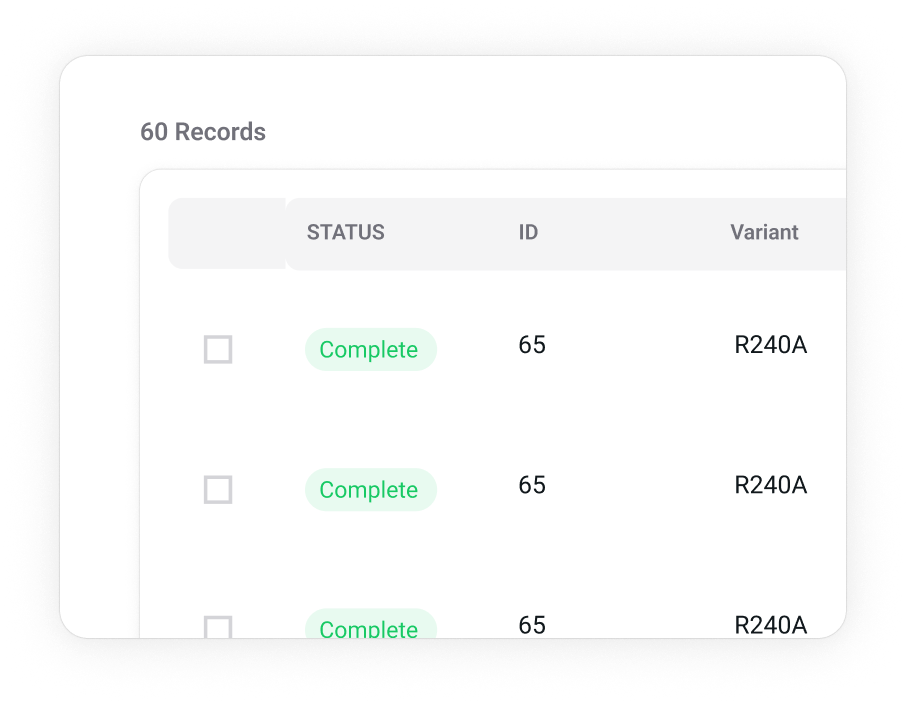
Characterize
The D2D system facilitates characterization contributions of thousands of students to solve the next generation challenge in protein design: function prediction.