About D2D Cure
Our Mission
The Design2Data Program involves students in an investigation of the sequence-structure-function relationship of proteins. Using the lens of protein biochemistry, students expand their knowledge and develop skills by engaging in a research workflow representative of cutting-edge biotechnology training in high demand by employers.
A feature that makes D2D unique among other multi-institutional protein biochemistry CUREs is its connection to the protein modeling research community, RosettaCommons, which uses the student-generated data to improve functionally predictive enzyme-design algorithms.
Large, uniformly obtained measurements of biophysical properties covering a broad and diverse sequence space across a wide range of enzymes are needed to truly begin utilizing the power of modern computational tools for creating predictive models of enzyme function.

Meet Our Enzymes
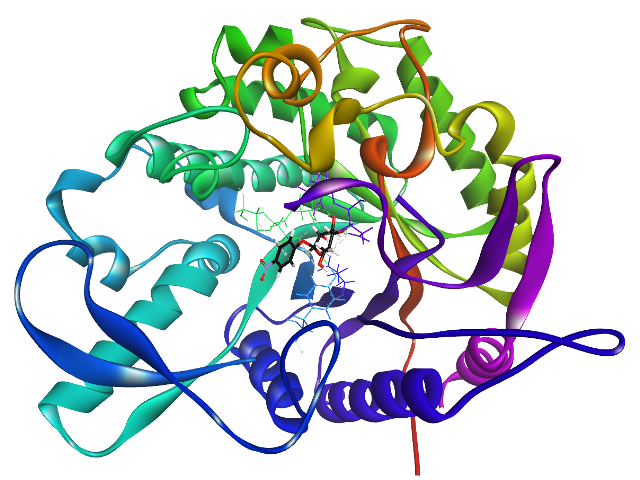
β-glucosidase B
An enzyme that catalyzes the hydrolysis of glucose monosaccharides from larger molecules at a β-glycosidic linkage
Learn More >
Future Data
We are actively seeking to identify more enzymes to expand our database.
Coming SoonMeet The Faculty
D2D Network Map
D2D Network Map shows geographical locations of nodes, lists faculty
instructors, includes course descriptions, and categorizes CURE variations
D2D LinkedIn Group
The goal of the D2D LinkedIn group is to facilitate networking and build a community of professionals around our shared D2D research project, fostering collaboration and knowledge-sharing among members!
Join our group

Special thanks to our technical team
This website was designed and developed by a group of students from , a software development and design agency at the University of California, Davis.
, a software development and design agency at the University of California, Davis.
Project Managers: Mohnish Gopi and Jess Fong
Developers: Vishal Koppuru, Ryan Uyeki, Hussain Ali, Kevin Bao, Vikram Karmarkar, Rahul Buhdiraja
Designers: Edyn Stepler, Samantha Tran
Learn more about CodeLab